Converting Datasets with file_tree
We often deal with structured imaging datasets, and understanding a different structure, or converting between them for data sharing or analysis can be tricky. I recently did this for a consolidated analysis and found that Michiel Cottaar’s file_tree module took much of the hassle out.
The file_tree.convert function can also handle making symlinks to save space and keep source data folders clean, and overwriting prevous runs.
As an approach I found this clear, flexible and powerful.
This has now been incorporated in file_tree (0.7.2 onwards) and a minimum working example is here:
https://gitlab.com/evan.edmond/filetree-convert-demo
First, we write a text file describing the structure of each dataset and the eventual desired target.
dataset 1:
HV_{participant} (subj_dir)
F3T_NNNN_NNN_{scancode} (scan_dir)
images_{image_n}_boldmbep2d2mmMB6v2RS.nii.gz (func_data_unclassified)
images_{image_n}_t1mprax1mmisowithNose32ch1001.nii.gz (T1w_unclassified)
sbref.nii.gz (func_sbref)
funcdata.nii.gz (func_data)
T1w.nii.gz (T1w)
twix_symlinks
slsr_csi_mcycle_dw_{block_n}.dat (mrsi_data)
dataset 2:
raw_data
C{participant} (subj_dir)
T1.nii.gz (T1w)
CSI
slsr_csi_mcycle_dw_{block_n}.dat (mrsi_data)
T1_dcm (T1w_dicom_dir)
rest
images_{image_n}_MB8FMRIfov21024mmresting.nii.gz (func_data_unclassified)
sbref.nii.gz (func_sbref)
funcdata.nii.gz (func_data)
fmap_rest
images_{image_n}_fieldmapgre2mmFoV216mm1001.nii.gz (fmap_mag)
images_{image_n}_fieldmapgre2mmFoV216mm2001.nii.gz (fmap_phase)
dataset 3:
raw
sub-{participant} (subj_dir)
ses-placebo (ses_dir)
anat
sub-{participant}_ses-p_T1w.nii.gz (T1w)
fmap
sub-{participant}_ses-p_magnitude1.nii.gz (fmap_mag)
sub-{participant}_ses-p_phasediff1.nii.gz (fmap_phase)
func
sub-{participant}_ses-p_task-{task}_bold.nii.gz (func_data)
sub-{participant}_ses-p_task-{task}_sbref.nii.gz (func_sbref)
mrsi
sub-{participant}_ses-p_slsr_csi_mcycle_pre_{block_n}.dat (mrsi_data)
Then a desired target dataset structure BIDS :
rawdata
sub-{participant} (input_subj_dir)
anat (input_anat_dir)
sub-{participant}_T1w.nii.gz (T1w)
sub-{participant}_T1w_mask.nii.gz (hand_area)
sub-{participant}_T1w_dicom (T1w_dicom_dir)
fmap (input_fmap_dir)
sub-{participant}_magnitude1.nii.gz (fmap_mag)
sub-{participant}_phasediff1.nii.gz (fmap_phase)
func (input_func_dir)
sub-{participant}_task-{task}_bold.nii.gz (func_data)
sub-{participant}_task-{task}_sbref.nii.gz (func_sbref)
mrsi (input_mrsi_dir)
sub-{participant}_vox-m1_csi_slaser_{block_n}.dat (mrsi_data)
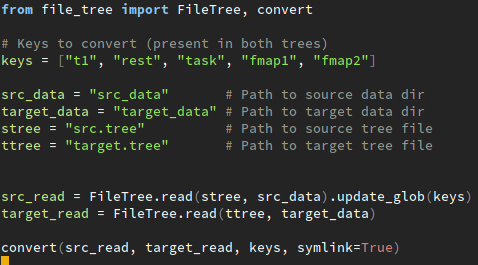
Conversion then can be done as follows:
from file_tree import FileTree, convert
# Keys to convert (present in both trees)
keys = ["t1", "rest", "task", "fmap1", "fmap2"]
src_data = "src_data" # Path to source data dir
target_data = "target_data" # Path to target data dir
stree = "src.tree" # Path to source tree file
ttree = "target.tree" # Path to target tree file
src_read = FileTree.read(stree, src_data).update_glob(keys)
target_read = FileTree.read(ttree, target_data)
convert(src_read, target_read, keys, symlink=True)